Cite As
We assessed these algorithms by investigating a dataset of 2546 trials from 54 subjects. The dataset can be retrieved from https://doi.org/10.5281/zenodo.5266492. Detailed results of this evaluation can be found in the manuscript
cite
Guggenberger et al. Evaluation of signal analysis algorithms for ipsilateral motor evoked potentials induced by transcranial magnetic stimulation (under review)
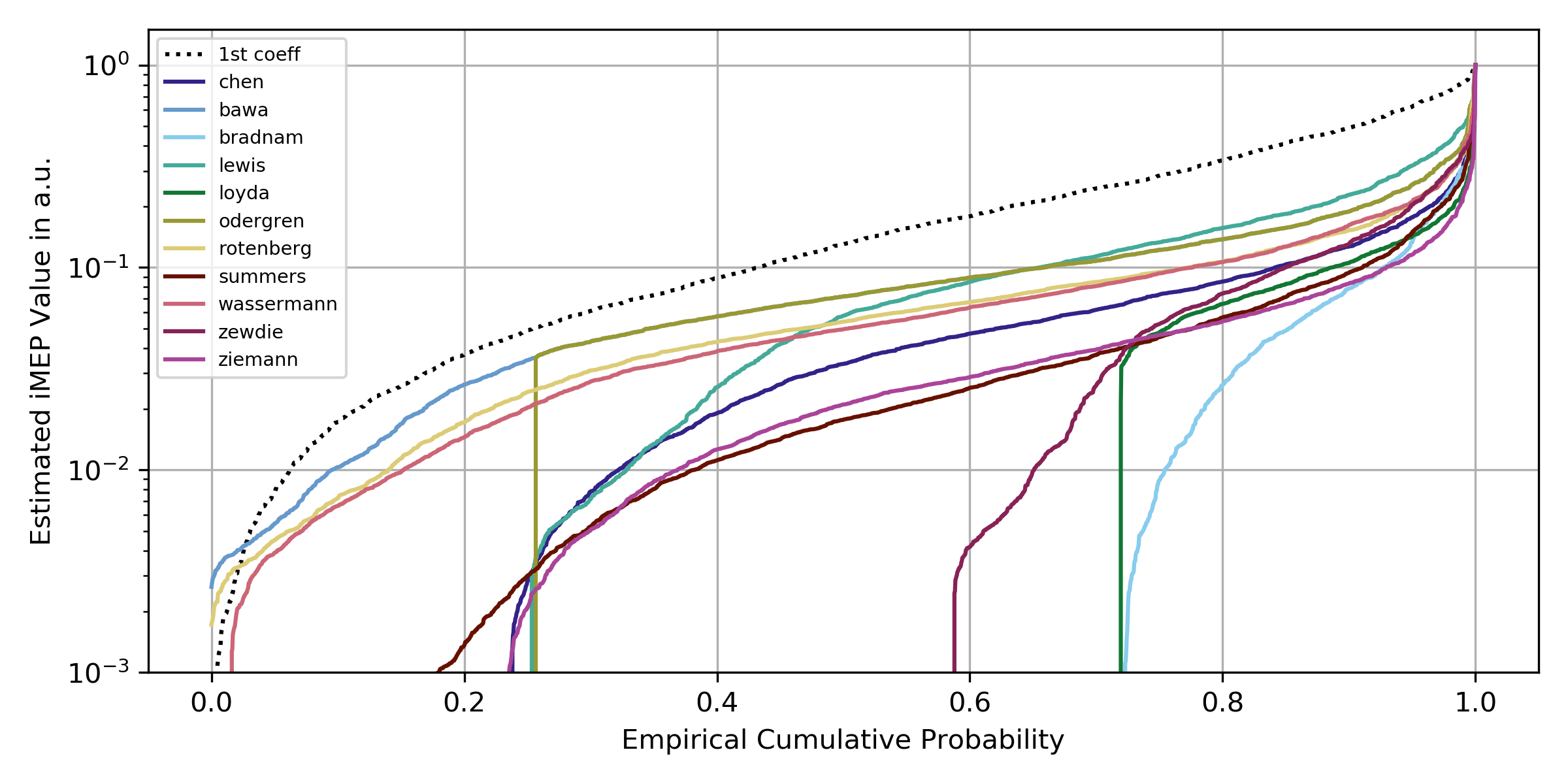
Strictness of algorithms
One of our findings showed that very strict threshold-based approaches, such as Bradnam or Loyda, are better able to recover the latent waveform, but that this comes at the cost of low reliability, as many trials cannot be measured and are classified as zero. This assignment of zeros can be interpreted as strictness, and can be estimated by plotting the empirical cumulative distribution function. The further to the right that an algorithm begins to assign values, the stricter it is. The level of strictness determines the number of trials that are discarded.
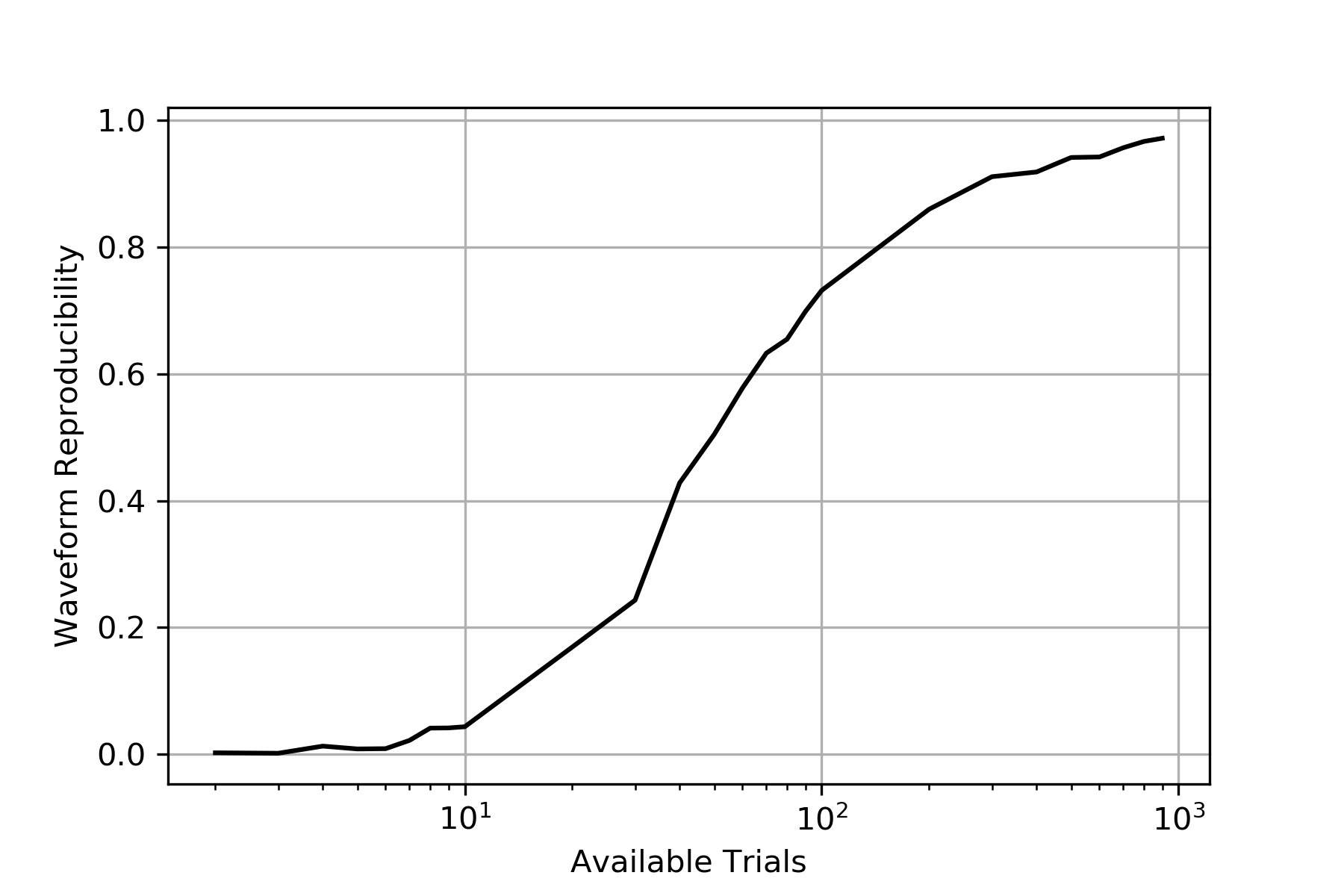
Number of trials required for decomposition
One alternative to template-based or threshold-based approaches is a direct decomposition, e.g., with a PCA. This opens the question, how many trials are sufficient to trust the results of a PCA?
To answer this question, we run a simulation. We picked at random two subsets with a specific number of trials from our dataset, calculated the two scores with PCA, and calculated Pearson’s correlation coefficient for them. This was performed over a range of sample sizes, and for each sample size, multiple repetitions were performed. See also Decomposition Simulation.
Visual inspection suggests that around 1000 trials would be required until the decomposition reaches saturation. That means we would need around 1000 trials to make sure that two scores are sufficiently similar in shape.